Introduction
A plasmid is a DNA molecule that is separate from, and can replicate independently of, the chromosomal DNA. They are double-stranded and, in many cases, circular. Plasmids usually occur naturally in bacteria, but are sometimes found in eukaryotic organisms
plasmids are often used to purify a specific sequence, since they can easily be purified away from the rest of the genome. For their use as vectors, and for molecular cloning, plasmids often need to be isolated.
There are several methods to isolate plasmid DNA from bacteria, the archetypes of which are the miniprep and the maxiprep/bulkprep. The former can be used to quickly find out whether the plasmid is correct in any of several bacterial clones. The yield is a small amount of impure plasmid DNA, which is sufficient for analysis by restriction digest and for some cloning techniques.
In the latter, much larger volumes of bacterial suspension are grown from which a maxi-prep can be performed. Essentially this is a scaled-up miniprep followed by additional purification. This results in relatively large amounts (several micrograms) of very pure plasmid DNA.
Extraction is an easy and quick way to purify DNA from a mixture of proteins, lipids and nucleic acids. It is important to keep in mind the purpose of this procedure while performing it. The reason for this procedure is to separate the plasmid DNA from its associated proteins so that further manipulations can be done to it. Enzymes added to purified DNA in vitro can have unhindered access to it. These enzymes might be used for restriction mapping, ligation, sequencing, or other procedures to modify the DNA. A poorly purified plasmid preparation will only be partially accessible to the enzymes and this will cause many headaches in these later steps. For this reason, special care must be taken to ensure a pure DNA preparation.
Objective
To determine the various effects that affect enzymatic activities.
Results:
OD230 = 0.427
OD260 = 0.572
OD280 = 0.309
Ratio (OD260/ OD280)
= (0.572 ÷ 0.309)
= 1.85
Ratio (OD260/ OD230)
= (0.572 ÷ 0.427)
= 1.34
OD260 = 0.572
DNA concentration (µg/mL)
= 50 µg/mL x OD260 x dilution factor
= 50 x 0.572 x 50
= 1430.0µg/mL
Total yield in 500µL sample
= DNA concentration x volume of sample in milliliters
= 1430.0 µg/mL x 0.50 mL
= 715.0 µg
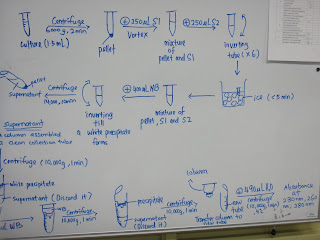 |
| Procedures |
 |
| Culture, S1,S2,Buffer NB,Wash buffer, Elution buffer. |
 |
| Centrifugal Machine |
 |
| Column-assembled collection tube |
 |
| Ice bath |
 |
| Centrifugation |
Disccussion:
From the calculation of result that we have got from the photospectrometer, the ratio of OD260 to OD280 was 1.85. The value is very close to 1.80. This shows that contamination of the protein is minimum.
Besides, the ratio of OD260/OD230 that we got was 1.34, which was smaller than 1.50. This reading represents the present of organic compound or chaotropic salts are present in the sample. This shown the amount of salt present in our sample were high because the greater the amount of salt, the lower the ratio.
Below are some interesting troubleshooting guides that we have found on the internet regarding extraction of plasmid DNA.
| Problem | Possible Reason | Solution |
| Poor bacterial growth | Inoculate bacterial cells from a plate or a cultural stock stored for long time | Always inoculate bacterial cells from a freshly streaked plate and grow with required antibiotics. |
| | Incubation with inadequate shaking | Grow cells with vigorous shaking (eg. 250rpm). Adjust a suitable shaking speed according to the angular magnitude of an orbital shaker platform. |
| Poor cell lysis | Use too many bacterial cells harvested from a large culture or over-grown culture. | Up to 5ml culture for high copy plasmid Up to 10ml culture for low-copy plasmid. When the culture is more than 5ml, use double amount of MX1, MX2 and MX3 buffer. |
| | Cell pellet is not well re-suspended | Do not add MX2 Buffer until cells are completely re-suspended by vortexing or pipetting. |
| Low yield of plasmid DNA | Not enough bacterial cells | Ensure that bacterial have grown well (OD600>1) after overnight incubation with vigorous shaking. |
| | Overgrowth of bacteria | Do not incubate for more than 16 hours. |
| | Plasmid does not propagate | Always inoculate bacterial cells from a freshly streaked plate and grow with required antibiotics. |
| | Insufficient or incomplete DNA elution | Efficient and complete DNA elution only take place when elution solution is within pH 7-8.5 and is in full contact with the membrane. Make sure that no less than 30µl of solution is dispensed onto the membrane and is completely adsorbed into it before centrifugation. |
| Low yield of plasmid DNA | Poor cell lysis | Refer to solution on Poor cell lysis |
| | Plasmid is larger than 10kb | Use elution solution preheated to 70cJ. |
| Plasmid appears smearing or degraded | Host strain is endA+ | Use endA- strain if possible. Or wash with WF buffer twice. Eluting DNA with TE buffer and storing at -20cJ can inhibit nuclease activity. |
| RNA contamination | No RNase A activity in MX1 Buffer | Ensure that RNase A is added into MX1 Buffer and stored at 4cJ |
| | Reduced Rnase A activity in MX1 Beffer due to improper or long-time storage | Add RNase A into MX1 Buffer to a concentration of 50µg/ml and store at 4cJ. |
| Plasmid DNA cannot be digested well or does not deposit into the well of gel during loading | Ethanol in WS Beffer is not completely removed | After washing with WS buffer, do discard the flow-through abd centrifuge the column at full speed for 3 minutes. If necessary, centrifugation for a few minutes more can completely remove ethanol. |
| Plasmid is denatured and migrates faster than supercoiled form during electrophoresis | Incubate in MX2 Buffer for too long time | After MX2 Buffer is added, do not incubate for more than 5 minutes. |
Conclusion:
Extraction of plasmid DNA is very important in many fields, but to do the extraction, many careful steps must be taken.
References
http://www.beckmangenomics.com/products/dna_purification_and_cleanup/agencourt_genfind_v2.html